Computational analysis and tool development for improving our understanding of mycobacterial genomes
We aim to provide better annotations of mycobacterial genomes that include non-coding regions of the genome. We develop tools to improve our knowledge of these non-coding elements and their role in host tropism and differences in adaptation to stress. We collaborate with experimental labs and produce testable hypotheses that can improve our knowledge of mycobacterial species biology.
Selected publications: Gibson, Stiens et al. (2022); Stiens et al. (2021); Gibson et al. (2021); Ozuna et al. (2019).
Regulation of gene expression in health and disease
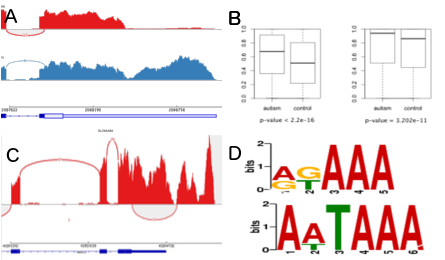
We focus on the computational study of gene expression using transcriptomic data and are particularly excited about the roles of non-coding RNAs in regulation of gene expression.
Selected publications: Szkop and Nobeli (2017); Szkop et al. (2017); Lee et al. (2017); Schwenk et al. (2017).
Protein-small molecule and protein-protein interactions
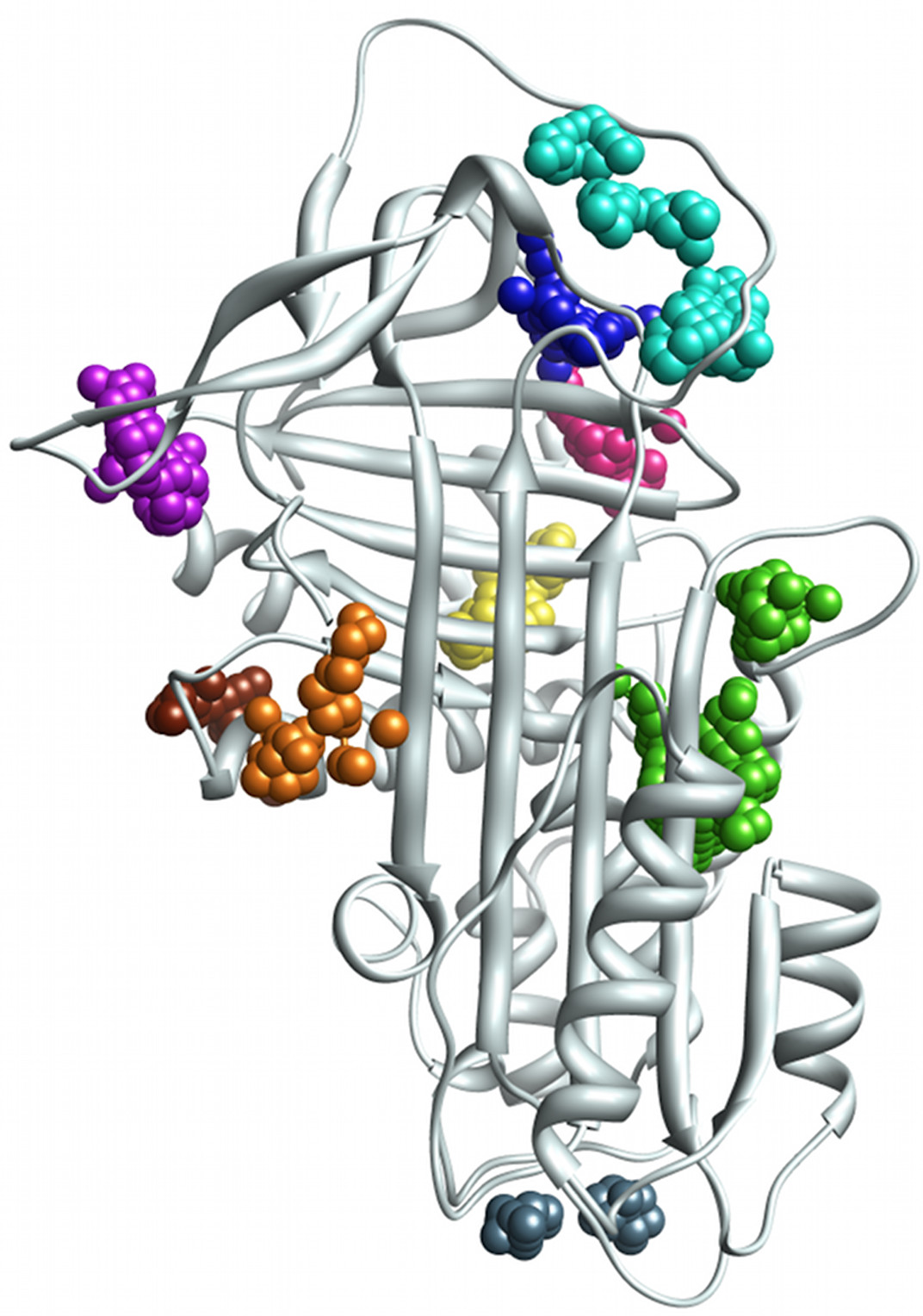
The group has a long history of applying computational methods (mainly docking) to identify promising leads in drug design projects, or ligands relevant to protein function. In the past, we have also explored ligand-binding and catalytic promiscuity and its evolutionary origins.
Selected publications: Proietti et al. (2016); Ashford et al. (2012); Patschull et al. (2012); Guzman et al. (2011); Favia et al. (2011); Gooptu et al. (2009); Nobeli et al. (2009); Macchiarulo et al. (2004).
The metabolome and metabolic pathways
We have a long-standing interest in the analysis of the collections of endogenous metabolites in model organisms from a structural and physicochemical point of view. We use chemoinformatics methods to reveal the relationships between metabolites within a species, compare metabolites from different species, and finally compare endogenous metabolites to exogenous (human made or environmental) small molecules.
Selected publications: Macchiarulo et al. (2009); Bashton et al. (2009); Bashton et al. (2006); Nobeli Thornton (2006); Nobeli et al. (2003).
