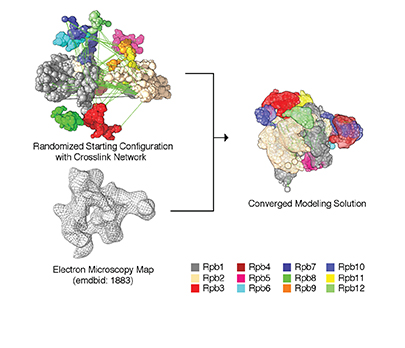
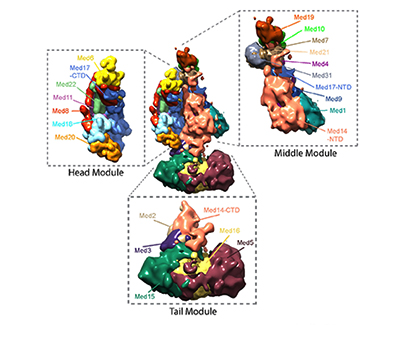
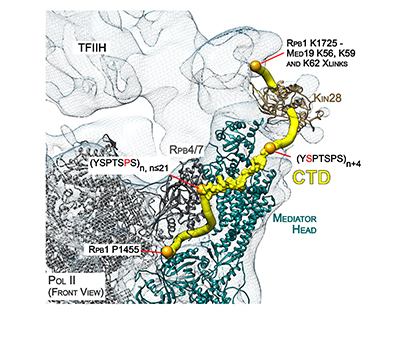
Overview
We are interested in gaining a molecular understanding of the processes that regulate DNA accessibility and control the transcriptional state of gene promoters. Eukaryotes depend on the activity of a collection of macromolecular machines to modulate chromatin structure and dynamics at gene regulatory elements. These machines are responsible for overcoming the repressive effects of chromatin packaging and provide DNA access to the basal transcriptional machinery. Chromatin remodelling complexes are a large superfamily of ATP-dependent DNA translocases that use the energy of ATP hydrolysis to modify the DNA-histone contacts within nucleosomes, driving nucleosome mobilisation or eviction. Mutations that disrupt the normal function of chromatin remodelling complexes have been implicated in a surprisingly large number of human neurodevelopmental disorders and tumour types.
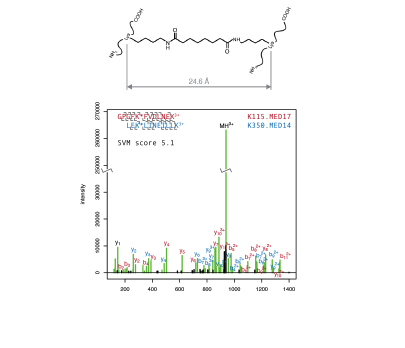
We use an integrative structural biology approach to investigate the molecular structure and mechanism of macromolecular chromatin modifying machines. In this approach we combine data from X-ray crystallography, cryo-electron microscopy, mass spectrometry, biophysical analysis and computational modelling strategies to build a detailed molecular picture of the 3-dimensional organisation and functional dynamics of these complicated multi-subunit protein assemblies.