Oct 2006 Chaperone lecture notes: 6 slides per page
2 slides per page
EM lecture notes: 6 slides per page (nb: 5.2 MB!)
2 slides per page (nb: 31 pages, 1.8 MB!)
Definition
A large group of unrelated protein families whose role is to
stabilize unfolded proteins, unfold them for translocation across
membranes or for degradation, and/ or to assist in their correct
folding and assembly.
Properties
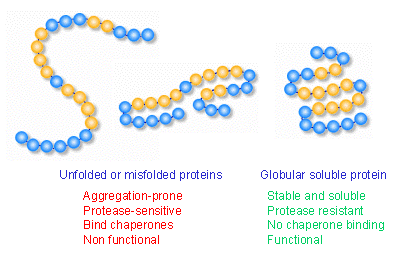
Main role: They prevent inappropriate association or aggregation of exposed hydrophobic surfaces and direct their substrates into productive folding, transport or degradation pathways.


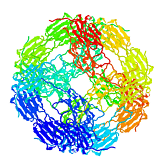
The structure of an archaebacterial small heat shock protein. It has 24 subunits, each with an immunoglobulin fold, arranged as a hollow shell with holes. It was determined by Kim et al (1998) Nature 394, 595-599, and is a member of the family that includes the eye lens protein alpha-crystallin. [pdb code 1shs]

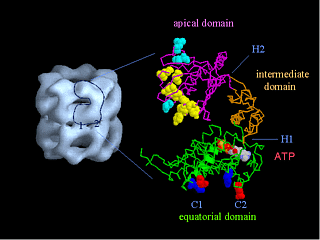
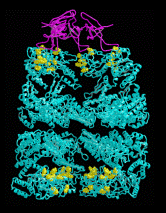
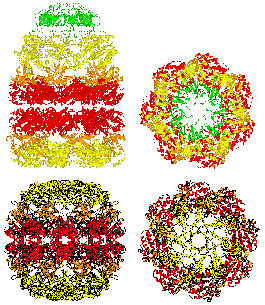
The structure of the chaperonin GroEL (hsp60) Left, a low resolution view of the 14-mer, from the X-ray crystal structure filtered to 25 A resolution. There are 2 contacts (numbered) between the two back-to-back heptameric rings. Right, a single subunit (60 kDa) shown as an alpha-carbon trace. There are three domains, separated by hinge regions (marked H1 and H2). Bound ATP is shown in space filling form, and the yellow residues are hydrophobic sites of substrate (non-native polypeptide) binding. These residues are also required for GroES (hsp10) binding, in addition to the blue residues. The charged residues in the inter-ring contacts are shown in red and blue. The structure was determined by Braig et al (1994) Nature 371, 578-586; Braig et al (1995) Nature Structural Biology 2, 1083-1094.
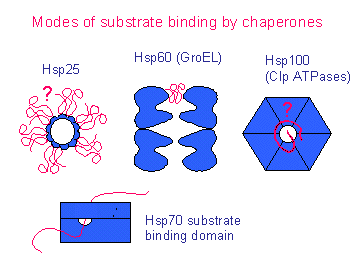
FAMILIES OF MOLECULAR CHAPERONES
Small heat shock proteins (hsp25) [holders]
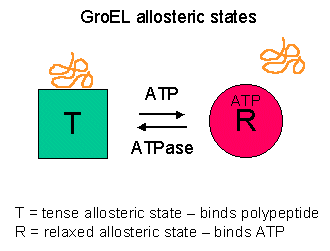
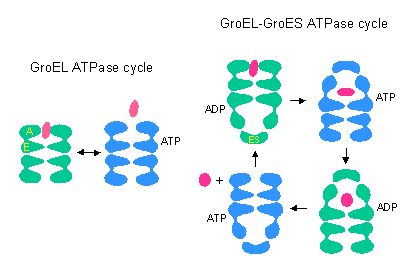
Hsp60 system (cpn60, GroEL) ATPase [(un)folders]
Hsp70 system (DnaK, BiP) ATPase [(un)folders]
Hsp90 ATPase [holder]
Hsp100 (Clp) ATPase [unfolder]
Calnexin, calreticulin
Folding catalysts: PDI, PPI [folders]
Prosequences: alpha-lytic protease, subtilisin (intramolecular chaperones) [folders]
|
|
||
|
Location |
Chaperone |
Roles |
|
HSP70 Family |
||
|
Prokaryotic cytosol |
DnaK
|
Stabilizes newly synthesised polypeptides and preserves folding competence; reactivates heat-denatured proteins; controls heat-shock response |
|
Eukaryotic cytosol |
SSA1, SSB1(yeast)
Hsc/hsp70, hsp40
(mammalian) |
Protein transport across organelle membranes; binds nascent polypeptides; dissociates clathrin from coated vesicles; promotes lysosomal degradation of cytosolic proteins |
|
ER |
KAR2, BiP/Grp78 |
Protein translocation into ER |
|
Mitochondria/ Chloroplasts |
SSC1
ctHsp70 |
Protein translocation into mitochondria; Insertion of light-harvesting complex into thylakoid membrane |
|
HSP60/CHAPERONIN Family GroE subfamily |
||
| Prokaryotic cytosol |
GroEL/ GroES |
Protein folding, including elongation factor, RNA polymerase. Required for phage assembly |
| Mitochondria/ Chloroplasts |
Hsp60/10 Cpn60/10 |
Folding and assembly of imported proteins |
|
TCP-1 subfamily |
||
| Archaebacterial cytosol |
TF55 Thermosome |
Binds heat-denatured proteins and prevents aggregation |
|
Eukaryotic cytosol |
TCP-1, CCT, or Tric |
Folding of actin and tubulin; folds firefly luciferase in vitro |
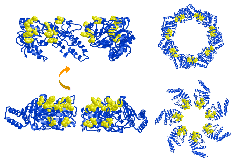
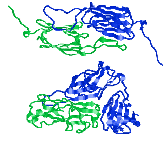
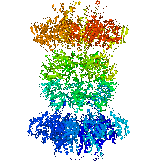
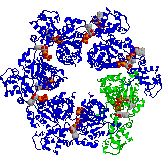 The end view of HslU with one subunit coloured in green, to show the inter-subunit
contacts. Each subunit has a bound ATP shown in space-filling format.
HslU is a member of the Hsp100 family, which forms part of the AAA ATPase superfamily. Many members of this superfamily have unwinding, unfolding or disassembly
functions.
The end view of HslU with one subunit coloured in green, to show the inter-subunit
contacts. Each subunit has a bound ATP shown in space-filling format.
HslU is a member of the Hsp100 family, which forms part of the AAA ATPase superfamily. Many members of this superfamily have unwinding, unfolding or disassembly
functions.
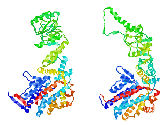
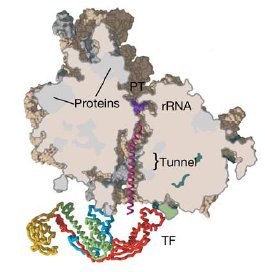 A section through the archaeal ribosome complexed with E. coli Trigger Factor (TF). The
polypeptide exit tunnel leads to an enclosed space cradled by TF, which is
proposed to allow folding of the nascent chain in an environment protected from
aggregation and proteolysis.
A section through the archaeal ribosome complexed with E. coli Trigger Factor (TF). The
polypeptide exit tunnel leads to an enclosed space cradled by TF, which is
proposed to allow folding of the nascent chain in an environment protected from
aggregation and proteolysis.
Molecular Chaperones: References
Reviews
Gething, M.J. & Sambrook, J. (1992) Protein folding in the cell. Nature 355,33-45.
Morimoto, R, Tissieres, A & Georgopoulos, C, Eds. (1994) The biology of heat shock proteins and molecular chaperones. Cold Spring Harbour Laboratory Press, Cold Spring Harbour, NY, USA.
Hartl, FU (1996) Molecular chaperones in cellular protein folding. Nature 381, 571-580.
Bukau & Horwich (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92, 351-366.
Sigler, PB, Xu, Z, Rye, H, Burston, SG, Fenton, WA & Horwich, AL (1998) Structure and function in GroEL-mediated protein folding. Ann. Rev. Biochem. 67, 581-608.
Ranson, N.A., White, H.E. & Saibil, H.R. (1998) Chaperonins. Biochem. J. 333, 233-242.
Saibil, H. (2000) Molecular chaperones: containers and surfaces for folding, stabilising or unfolding proteins. Current Opinion in Struct. Biol. 10, 251-258.
Molecular chaperones - review volume - Advances in Protein Chemistry, vol 59 (2002).
Saibil, H & Ranson, N. (2002) The chaperonin molecular machine, Trends in Biochem. Sci. 27, 627-632.
Sakahira,H, Breuer, P, Hayer-Hartl, Mk, Hartl, FU (2002) Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. PNAS 99, 16412-16418.
Bukau, B, Weissman, J, Horwich A (2006) Molecular chaperones and protein quality control. Cell 125, 443-451.
Research Papers
Anfinsen, C.B. (1973) Principles that govern the folding of protein chains. Science 181, 223-230.
Zhu, X, Zhao, X, Burkholder, W, Gragerov, A, Ogata, C, Gottesman, M & Hendrickson, W (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606-1614.
Braig, K., Otwinowski, Z., Hegde, R., Boisvert, D.C., Joachimiak, A., Horwich, A.L. and Sigler, P.B. (1994). The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature 371, 578-586.
Fenton, W.A., Kashi, Y., Furtak, K. & Horwich, A.L. (1994) Residues in chaperonin GroEL required for polypeptide binding and release Nature 371, 614-619.
Roseman, A, Chen, S, White, H, Braig, K & Saibil, H (1996) The chaperonin ATPase cycle: Mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell 87, 241-251.
Buckle, A., Zahn, R. & Fersht, A. (1997) A structural model for GroEL-polypeptide recognition. Proc. Natl. Acad. Sci. USA 94, 3571-3575.
Xu, Z., Horwich, A. & Sigler, P. (1997) The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388, 741-750.
Ditzel et al (1998) The crystal structure of the thermosome. Cell 93, 125-138.
Kim, Kim & Kim (1998) Crystal structure of a small heat shock protein. Nature 394, 595-599.
Rutherford SL, Lindquist S. (1998) Hsp90 as a capacitor for morphological evolution. Nature 396, 336-342.
MC Sousa, CB Trame, H Tsuruta, SM Wilbanks, VS Reddy, and DB McKay (2000) Crystal and Solution Structures of an HslUV Protease-Chaperone Complex.Cell 103, 633-643.
Ferbitz et al (2004) Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431, 590-596.
Structural basis of interdomain communication in the Hsc70 chaperone. Jiang J, Prasad K, Lafer EM, Sousa R. (2005) Mol Cell. 20, 513-524.
Bukau, B, Weissman, J, Horwich A (2006) Molecular chaperones and protein quality control. Cell 125, 443-451.
Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. (2006) Nature 440, 1013-1017.
Structure of an Hsp90-Cdc37-Cdk4 complex. Vaughan, CK, Gohlke, U, Sobott, F, Good, VM, Ali, MMU, Prodromou, C, Robinson, CV, Saibil, HR & Pearl, LH (2006) Mol. Cell 23, 697-707.