Bacterial Toxins
Epsilon-toxin
We have taken an important step forward in understanding another of the major toxins
produced by Clostridium perfringens. Epsilon-toxin is produced by
strains of the bacterium that inhabit the gut of sheep and lambs.
Intoxication results in enterotoxaemia and neurological disorders and
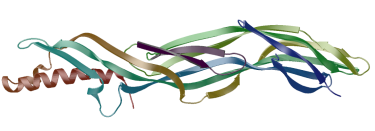
is usually fatal. We have solved the structure of the prototoxin by X-ray analyis.
The structure suggests that that it is a pore forming toxin
that changes conformation between the soluble and transmembrane states.
We have prepared a heptameric form of the protein that is probably involved
in pore formation and are studying this form by X-ray diffraction and electron
microscopy.
X-ray structure of epsilon-prototoxin
Nature Structural and Molecular Biology, (2004), 11, 797-798.
|
|
Gas-gangrene toxin
This enzyme, also known as alpha-toxin, is a phospholipase-C and is the key virulence determinant in
gas-gangrene. Gangrene is usually caused by an infection of a wounded limb by Clostridium perfringens and
although the bacterium is susceptible to drugs, the rapid destruction of muscle cells
by the secreted toxin usually leads to the need to amputate the infected limb.
Cell death is caused by the hydrolysis of the phospholipids of
cell membranes and the triggering of secondary messenger pathways.
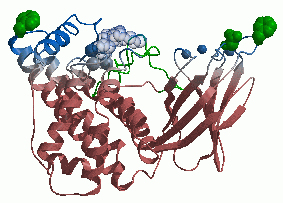
We have already solved crystal structures of the toxin. Most show three metal ions in the active site of
the enzyme in an open conformation which we believe to be catalytically active.
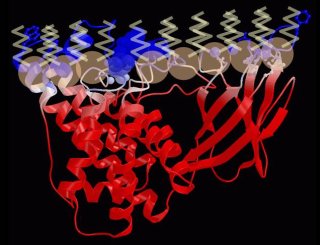
In ref (i) where we proposed a model for the interaction of the toxin with the cell membrane.
We are working with Professor Rick Titball
of the University of Exeter
to establish the mechanism of toxin-membrane interactions with the longer term goal of designing toxin inhibitors.
We wish to answer the following questions:
- What aspects of phospholipids and cell membranes particularly favour toxin binding?
- Which residues in the toxin are essential to membrane binding and do these residues
show conformational flexibility on binding membrane?
- What mechanism type does the toxin exhibit when cleaving membrane phospholipid?
- How does this explain the toxicity of the toxin in contrast to related bacterial phospholipase-Cs?
X-ray structure of gas-gangrene toxin
Nature Structural Biology, (1998), 5, 738-746.
|
|
Toxin-membrane interaction model
|
|
Entero-toxin
We have also conducted pilot studies on entero-toxin, a major cause of human food poisoning.
Superantigen-like proteins
Claire has also solved the structure of a bacterial protein that is a member of a family that are structually
analogous to some superantigens and used to be called SET proteins. More information is here.
References
(i) Clostridium perfringens epsilon-toxin shows structural similarity with the
pore forming toxin aerolysin, Cole, A, Gibert, M, Popoff, M, Moss, D,
Titball, R and Basak, A, Nature Structural and Molecular Biology, (2004), 11,
797-798.
(ii) Crystal structure of C. perfringens alpha-toxin with the active site closed
by a flexible loop region, Eaton, J T, Naylor C E, Howells A, Moss D S,
Titball, R W and Basak, A K, J. Mol. Biol. (2002), 319, 275-281.
(iii) The first strain of Clostridium perfringens isolated from an avian
source has an alpha-toxin with divergent structural and kinetic properties,
Justin N, Walker, N, Bullifent, H L, Songer G, Bueschel, D M, Jost H,
Naylor C, Miller, J, Moss, D S, Titball, R W and Basak, A K,
Biochemistry,(2002), 41, 6253-6262.
(iv) Mapping critical residues for toxicity in Clostridium perfringens
phospholipase C,
the key toxin in Gas gangrene: N Walker, J Holley, C Naylor, M Flores-Diaz,
A Alape-Giron, M Thelestam, D Moss, A Basak, J Miller and R Titball,
Arch. Biochem. Biophys., (2001), 384, 24-30.
(v) Structure of the key toxin in gas-gangrene, Naylor, C E, Eaton, J T,
Howells, A, Justin, N, Moss, D S, Titball, R W and Basak, A K, Nature
Structural Biology, (1998), 5, 738-746.
Any questions and bug reports to
David Moss
Last updated 8 January 2014
d.moss@mail.cryst.bbk.ac.uk



