Amyloid Assemblies
Crystallography, ISMB
Birkbeck College London
Amyloid Assemblies |
 |
|
Helen Saibil
Jonathan O'Driscoll Former group members Silvia Panico (Imperial College) Lilia Milanesi (Queen Mary College) Jose Jimenez (Sanger Centre) Julie Hodgkinson (Medizinische Hochschule Hannover) Sara Cohen-Krausz made important contributions to our amyloid work, and sadly lost her battle with cancer in May 2009. After her stay at Birkbeck, she worked in the Hebrew University, Jerusalem, until her untimely death. |
|
Amyloid fibrils are insoluble aggregates that result from the self-assembly of partially unfolded proteins. Regardless of the native structure of the precursor proteins, the predominant secondary structure in the fibrillar form is beta sheet. Proteins that form amyloid in vivo are associated with diseases such as Alzheimer's and CJD.
SH3 Amyloid (Jimenez et al.) |
| In collaboration with Dr Margaret Sunde (now at University of Sydney, Australia) and Prof Chris Dobson , we determined the low resolution structure of amyloid fibrils formed from a SH3 domain by cryo-electron microscopy and image processing (Jimenez, J.L., Guijarro, J.I., Orlova, E., Zurdo, J., Dobson, C.M., Sunde, M. and Saibil, H. (1999) Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J 18, 815-821). Further work was published in: Dependence on solution conditions of aggregation and amyloid formation by an SH3 domain. Zurdo J, Guijarro JI, Jimenez JL, Saibil HR & Dobson CM (2001) J Mol Biol. 311, 325-340. |
Insulin amyloid fibrils (Jimenez et al.)The polypeptide hormone insulin consists of two chains linked by disulfide bonds. The native structure is almost completely alpha-helical, but denatured insulin readily forms amyloid fibrils, which requires a complete refolding of the insulin chains. In collaboration with Professors Chris Dobson and Carol Robinson, University of Cambridge, we have used cryo EM to characterise several different forms of insulin amyloid fibrils (The protofilament structure of insulin amyloid fibrils. Jimenez, JL, Nettleton, E, Bouchard, M, Dobson, CM, Robinson, CV & Saibil, HR (2002) Proc. Natl. Acad. Sci. USA 99, 196-201). |
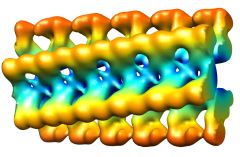
Ex vivo lysozyme fibrils (Jimenez et al.)In collaboration with Prof. Mark Pepys and Dr Glenys Tennent at the National Amyloidosis Centre, Royal Free and University College Medical School, we have examined lysozyme fibrils isolated from pathological tissue. The fibrils have a wavy shape. An averaged repeat from cryo EM images and a schematic 3D model in side view and an end view of the spiral model with 6 protofilaments are shown below. (Structural diversity of ex vivo amyloid fibrils studied by cryo-electron microscopy. (2001) Jimenez, JL, Tennent, G, Pepys, M & Saibil, HR, J. Mol. Biol. 311, 241-247). |
 |
  |
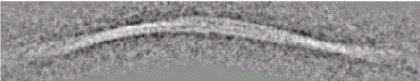
Mammalian Prion fibrils (Tattum et al.)In collaboration with Profs. John Collinge and Tony Clarke at the MRC Prion Unit, Institute of Neurology, University College London we have examined PrP fibrils formed in vitro. |

| Elongated oligomers assemble into mammalian PrP amyloid fibrils. Tattum et al (2006) J Mol Biol 357, 975-985. |
Beta-2 microglobulin amyloidIn collaboration with Prof. Sheena Radford at Leeds University we are studying the 3D structure and properties of beta-2 microglobulin amyloid fibrils. |

| Globular tetramers of beta-2 microglobulin assemble into elaborate amyloid fibrils. White et al (2009) J Mol Biol 389, 48-57. |
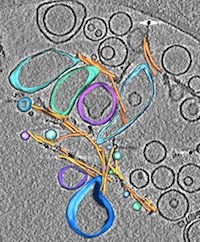 |
Direct three-dimensional visualization of membrane disruption by amyloid fibrils. Milanesi et al PNAS (2012) |
Yeast PrionsIn collaboration with Prof. Achilleas Frangakis at University of Frankfurt and Jens Tyedmers at ZMBH, Heidelberg we are studying yeast prion assemblies. This work was started during a sabbatical at EMBL Heidelberg. |
 |
Heritable yeast prions have a highly organized three-dimensional architecture with interfiber structures. Saibil et al PNAS 109, 14906-14911 (2012) |
|
EM group web page Helen Saibil |